
Insights from Professor Wilhelm Huck
Over the past decade, droplet microfluidics has clearly proven its value. Droplet-based technologies provide a high-throughput and sensitive method to perform quantitative screening assays by encapsulating single cells in picoliter compartments. This single cell approach is transforming our understanding of complex biological processes at the cellular and molecular level, and the continuous advancements in this field are fascinating.
To learn more, we spoke with Wilhelm Huck, Professor of Physical Organic Chemistry at the Institute for Molecules and Materials, Radboud University Nijmegen, and a co-founder and Scientific Advisory Board member of Sphere Fluidics. In this article, Professor Huck discusses the application of single cell analysis in diagnostics and his work using droplet-based microfluidics to detect and isolate circulating tumor cells (CTCs). We also explore the future potential of single cell technologies and new developments taking place at Sphere Fluidics.
Single cell analysis using droplet microfluidic technologies
My research in microfluidics goes back to my postdoc in 1997-99. At first, I was using continuous-flow based systems. Shortly after, the field diversified with the emergence of droplet microfluidics, where two immiscible liquids are forced through microfluidic channels to generate monodisperse pico- to nanoliter water-in-oil droplets. I began conducting research using these technologies at Cambridge University, alongside Chris Abell, Florian Hollfelder, and Andrew Griffiths. Back in 2004-2005, the original aim was to use droplets as a chemical factory where each droplet would produce a different enzyme to select for a different chemical reaction.
In the past 5 years, the study of compartmentalized single cell has led to a revolution in biology. One advantage of droplet microfluidics is the ability to work with tiny volumes, so even though you are studying a single encapsulated cell, the contents of the droplet are still in reasonably high concentration. Combine the fact that large numbers of droplets can be interrogated individually, together with the fact that cell encapsulation enables further analysis of genetic materials and proteins, and it is clear that droplet microfluidics make it possible to explore the properties of large numbers of individual cells.
Breaking new ground in single cell diagnostics
Within diagnostics, many different directions can be pursued with single cell analysis. One notable application is the isolation and detection of rare cells from a large population of other cell types, such as screening and detecting cancer biomarkers, including CTCs that exist in minute quantities (1 to 10 cells per 10 ml of blood). By encapsulating single cells in picodroplets, each cell can be examined individually without non-specific noise coming from the other cells in the background. The picodroplet format allows for higher-throughput and higher-sensitivity assays, making a big difference in diagnosis. Other expanding application areas include single cell transcriptomics, single cell proteomics, and single cell genomics.
Detecting Circulating Tumor Cells
In a recent study, we have utilized a single cell droplet microfluidic technique to detect and count CTCs in blood. Circulating tumor cells are an example of extremely rare cells that dislodge from the primary tumor, travel through the bloodstream, and settle somewhere else to form a secondary tumor. There is substantial evidence that the number of these CTCs in your blood (per millimeter of blood) correlates with the severity of the metastasis and poor prognosis of the patient. Improving the detection of CTCs can aid in early diagnosis and improve cancer treatment.
Finding CTCs has been typically very difficult. One existing method involves using the cell’s unique features and recognizing specific cell-surface markers such as Epcam (a cell surface protein), then separating these cells from the blood using magnetic beads with antibodies capable of detecting EpCAM. However, using this procedure, you only find those cells that are EpCAM-positive, and all other CTCs are not detected.
Alternative methods are based on the mechanical properties of CTCs and use microfluidic devices to ‘sieve’ or sort out tumour cells from healthy white blood cells. These methods work reasonably well, and there are commercial examples of that. However, the disadvantage is that the difference in the physical properties between those few CTCs and the millions of white blood cells are not enough to isolate only the CTCs. Therefore, there is a need to establish alternate approaches capable of improving the identification of CTCs that addresses the drawbacks of the current methods.
A droplet-based microfluidic approach
The approach presented in our research is based on the inherent metabolic properties of the cell. Since the 1920s, it has been known that tumors are more acidic than healthy tissue. While every cell in your body does secrete acid (usually, when starved of oxygen, cells secrete lactic acid), there is a marked difference in secretion rate between the tumor cells and your healthy white blood cells. We hypothesized that if we could measure the acid secretion of individual cells, we could detect tumor cells circulating in the blood.
This metabolic-based approach proved to be a simple and effective method for detecting and counting CTCs. First, we encapsulated and incubated individual cells in droplets, then screened for a change in pH of the droplets. Although one tumor cell will not change the pH of a milliliter of blood, when isolated in a picoliter droplet, enough acid is secreted to alter the pH and hence allow the tumor cell to be detected. By studying the number of acid-positive droplets in emulsified samples taken from prostate cancer patients, we demonstrated the prognostic value of CTCs. Whenever we detected more than five of these cells per 7.5 milliliter of blood, the life expectancy of the patients was low. Considering that a milliliter of blood has a billion red blood cells and 1-10 million white blood cells, it is quite scary how just a few CTCs can result in severe illness.
As part of the research, we also isolated acidic droplets and retrieved cells for single cell transcriptomics to understand what these cells are and are not. Surprisingly, we discovered that 90% of the acidic cells are not cancer cells; they are cancer-associated stromal cells. Our findings indicate that while tumor cells travel to other areas in the body, they do not survive for very long on their own. Instead, the stromal cells, such as cancer-associated fibroblasts, are hypothesized to seed new microniches for metastasis.
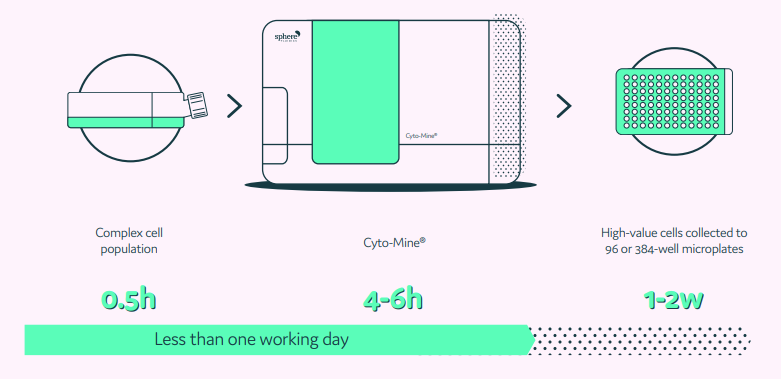
An integrated and fully automated microfluidic process using Cyto-Mine®
Future Research
In further studies, we will look at the treatment response of patients. In doing so, we will study the spectrum of cells to detect early signs of resistance against chemotherapy, because as cells change and the patient becomes insensitive to treatment, we may identify cell signatures associated with the acquired resistance. Additionally, cell heterogeneity might indicate the location of the metastasis as the cells may have a signature associated with prostate, lung, bone, or breast tissue. In a clinical setting, this information will inform doctors on the best course of treatment, as they will know where the CTCs are coming from and what the cells are carrying. Other therapeutic benefits of this kind method include antibody detection and cytokine detection.
An additional area we will investigate is using Sphere Fluidics’ Cyto-Mine® to optimize the analysis process, and especially extract individual acid-secreting cells from the sample. The fully integrated workflow of Cyto-Mine® would help to automate cell encapsulation, incubation, and detection workflow. At the moment, the droplet sizes we use are slightly different, but this can be adapted in Cyto-Mine®, as can the fluorescent dyes in the droplets.
Optimizing single cell analysis workflows
Single cell analysis typically requires a complex workflow encompassing cell line preparation, microfluidics, detection technologies, algorithms to analyse and interpret the data generated. Research labs have, therefore, needed to invest in multiple instruments and complicated processes that require specialist expertise. Using these processes, it is challenging to go beyond proof of principle experiments because it is often too difficult and costly to perform these experiments routinely every day. To simplify this process, increased integration and automation of the workflow is needed.
What Sphere Fluidics has achieved is the development of an integrated and automated single cell analysis system, Cyto-Mine®. Cyto-Mine® enables the high throughput encapsulation, screening, sorting, and dispensing of target cells into microtiter plates in just one day. The workflow is streamlined into one machine, and such integration is a huge step forward.
Ongoing and future developments at Sphere Fluidics
In the current research environment, we have a tremendous opportunity to help other organizations remove traditional bottlenecks and achieve improved throughput and reproducibility. The encapsulation of single cells in picodroplets provides a method to analyse single cells without interference from other cells and addresses the challenge of inefficient and irreproducible results. Additionally, Cyto-Mine® is a flexible platform offering high viability and assay sensitivity across a range of cell types such as B cells, hybridomas and CHO cells.
Following the development of the Cyto-Mine® single cell analysis platform, we continue to focus on expanding the system’s capabilities to deliver more for our clients. One of our immediate next steps is increasing the number of fluorescent excitation and detection options with the machine. Then beyond that, our other aspiration is to develop new application areas such as cell therapy research, genome editing and synthetic biology. Cyto-Mine® provides a way of detecting all kinds of cells that secrete different substances, whether metabolites, cytokines or antibodies, while also studying what is going inside the cell or even on the cell surface. Such a detection range is a tremendous opportunity that we can explore further.
About Sphere Fluidics
Sphere Fluidics’ platforms are built on over a decade of scientific and engineering research and development. Originally spun out from the University of Cambridge, we specialize in multiple technical areas to deliver miniaturization.
For further information contact:




